Genomic Sequencing for Wastewater-Based Epidemiology

Local outbreak detection and tracking is critical to the public health response to emerging or reemerging pathogens. Wastewater pathogen tracking is a valuable new approach that complements and enhances traditional pathogen surveillance and can feed new data into disease forecasting and predictive modeling to help communities make targeted policy decisions to prevent or control outbreaks.
Battelle is at the forefront of this developing technology. We’re establishing and optimizing the parameters and strategies of wastewater pathogen tracking so that this new data source can enable policymakers to understand how outbreaks are unfolding over time, which communities are affected, and which populations are most vulnerable. Our team has decades of experience examining chemical and biological agents in a variety of environments, including wastewater, to identify narcotics, opiates, influenza, SARS-CoV-2, and other hazards to human health. The information and analytical tools we provide help you proactively protect your community against pathogens, such as bacteria and viruses, and substances that are a threat to public health.
How does it work?
Shed pathogens flow into the wastewater from individuals who are infected or using these substances. Through methods to collect and analyze the samples we can detect the nucleic acids from all pathogens present in the wastewater to monitor what diseases are circulating in a community, if they are antibiotic resistant organisms, and the amount present.
Sampling
Households, businesses, and high priority locations like hospitals and nursing homes using sewer systems produce wastewater streams that ultimately reach local wastewater treatment plants. Samples of wastewater can be collected using automatic sampling devices at sewer service manholes or at wastewater treatment facilities. Metadata such as location, temperature, pH, type of sewer system, weather, and time can be tracked to provide valuable data that help not only detect pathogens, but also determine the spread and severity of an outbreak within a location or region over time to enable disease forecasting.
Analysis
Samples can be analyzed in multiple ways to detect and characterize pathogens including detection of new mutations or variants that may elevate the risk of outbreak spread. Samples are processed and tested with a combination of technologies such as quantitative PCR (qPCR), targeted nucleic acid sequencing, and untargeted metatranscriptomics (total RNA sequencing). In addition to these laboratory-based analyses, other data such as location, temperature, pH, sewer type, weather, and demographic variables of the community are incorporated. This information can be used to determine if vaccine or diagnostics need to be adapted to ensure continued efficacy and can help with supply chain planning.
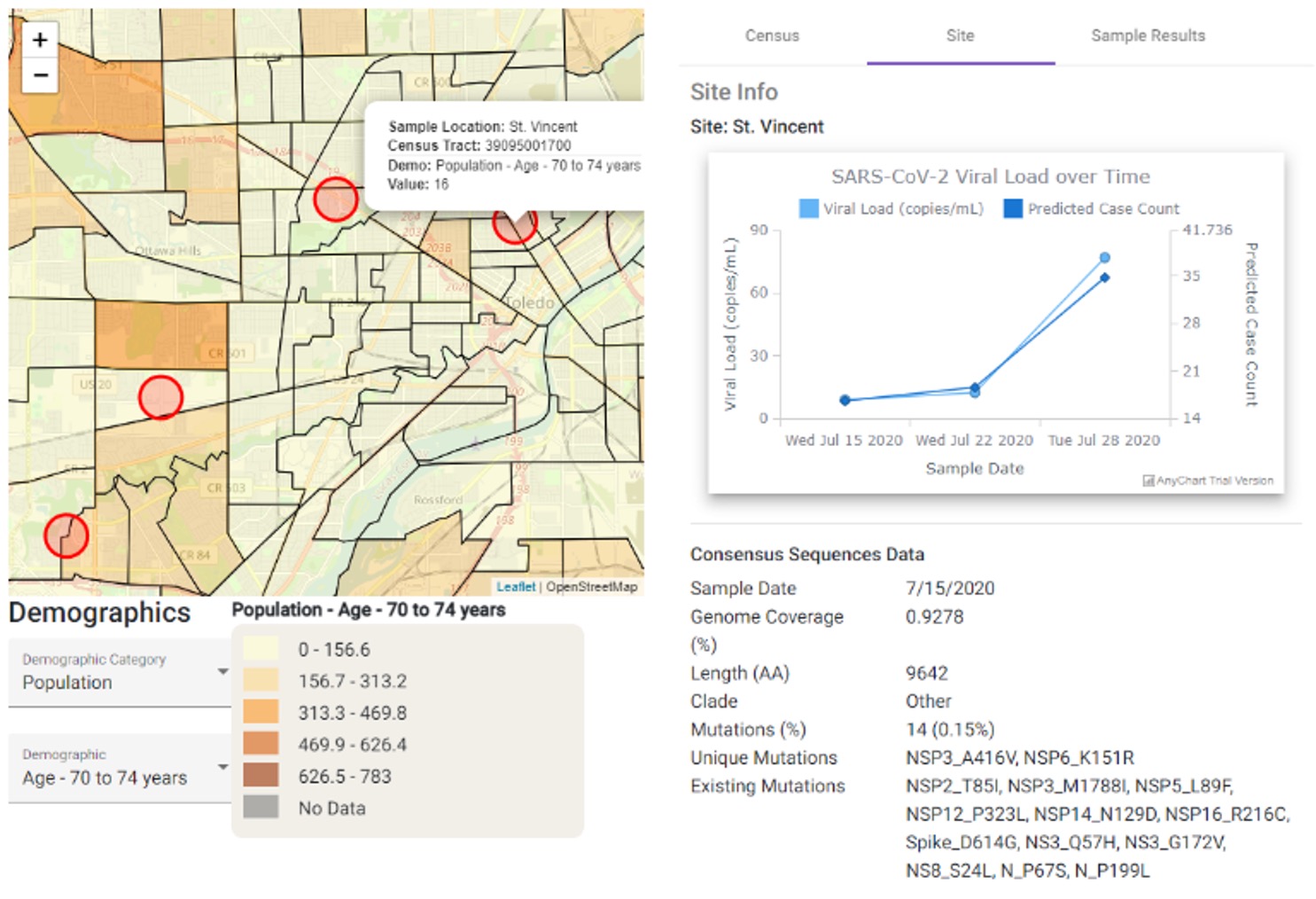
Communication and Data Visualization
Collected data is managed, analyzed, and communicated to stakeholders. Battelle’s Wastewater Pathogen Tracking Dashboard (WPTD) is a biosurveillance tool that tracks the incidence and spread of pathogens quantified through wastewater analysis. The tool incorporates both qPCR and sequencing data of wastewater samples collected in the region of interest, allowing users to view information at both the site and regional levels. Site-specific information includes data about the wastewater samples, summaries of sequencing results, and the ability to view those results at different time points. At the regional level, the WPTD provides an overlay of demographic variables of interest at a census tract resolution, allowing the user to understand the pathogen detections and prevalence in the context of the populations being affected. This dashboard provides a complete contextual picture that can be evaluated so that timely, data-informed decisions can be made to protect communities.
Related Blogs
BATTELLE UPDATES
Receive updates from Battelle for an all-access pass to the incredible work of Battelle researchers.


